This Fox Chase professor participates in the Undergraduate Summer Research Fellowship.
Learn more about Research Volunteering.
Related Articles
00 / 00

This Fox Chase professor participates in the Undergraduate Summer Research Fellowship.
Learn more about Research Volunteering.
Your Gift Will Fund Groundbreaking Cancer Epigenetics Research in the Whetstine Laboratory
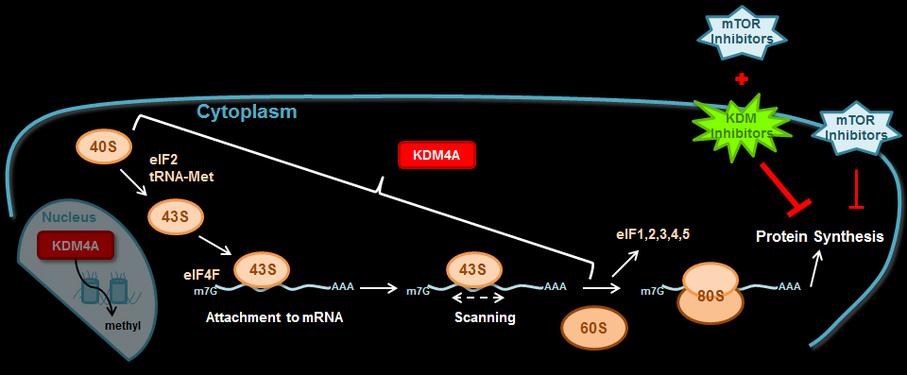
We are determining whether other chromatin factors enhance mTOR inhibitor sensitivity or other cancer therapies.
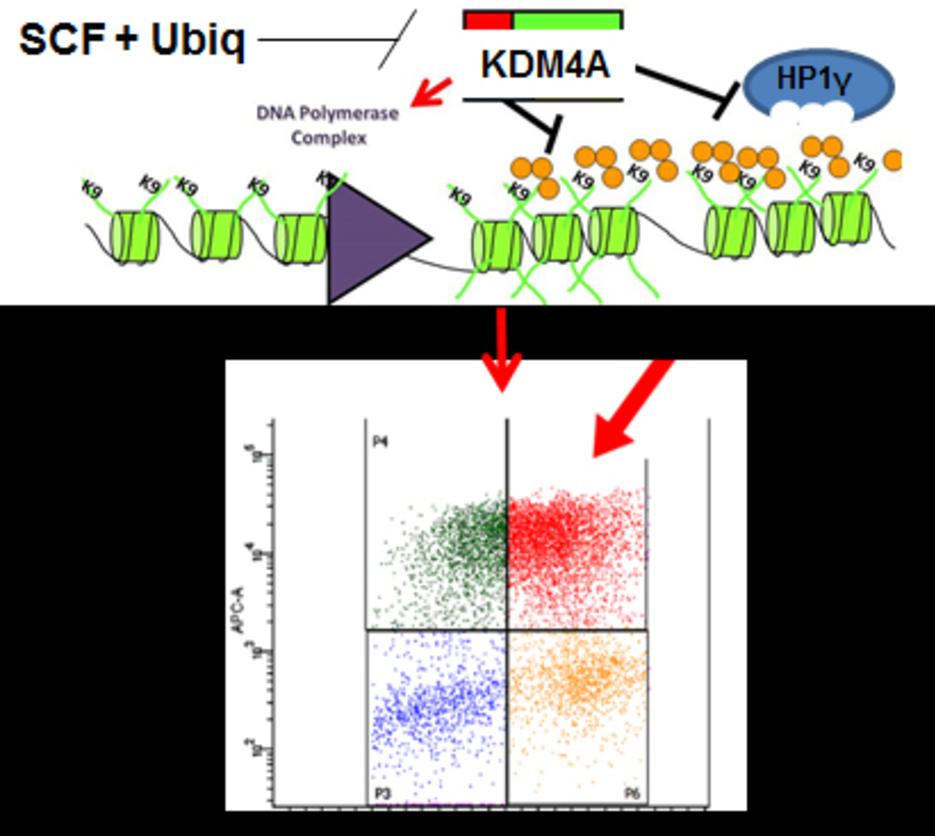
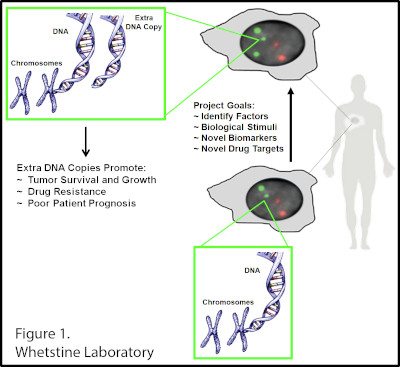
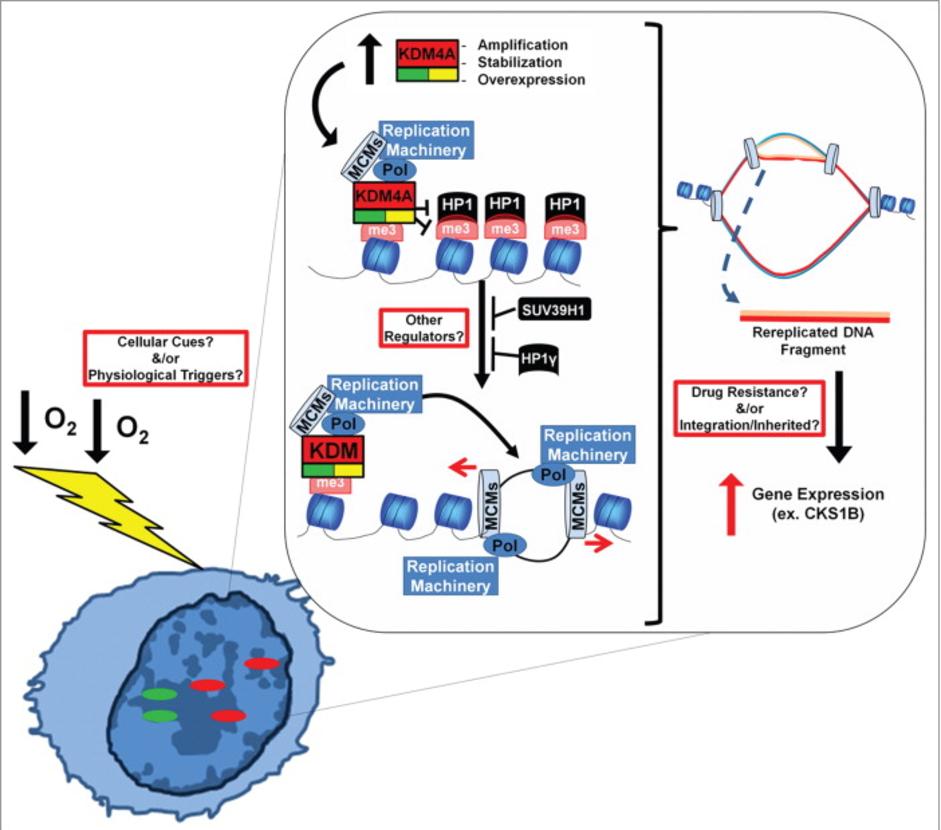
The Whetstine laboratory studies how chromatin factors control gene expression and genome stability. These types of studies were initiated by studying a specific class of chromatin regulators, JmjC-containing histone demethylases, that Dr. Whetstine was involved in discovering. While interrogating demethylase function, the Whetstine laboratory uncovered the first enzyme (KDM4A- histone tri-demethylase) responsible for generating extrachromosomal transient site-specific copy gains (TSSGs) of drug resistant oncogenes. Dr. Whetstine’s team is further interrogating these same relationships with other chromatin/epigenetic modifiers and DNA amplification events occurring in cancer and other diseases. His laboratory aims to provide mechanistic insights into extrachromosomal DNA amplification generation and the associated heterogeneity within cell populations, while identifying biomarkers to both predict and target copy gain events driving human diseases (Figure 1). The Whetstine laboratory is addressing the genomic features (epigenome and 3D genome organization) that are responsible for increasing the propensity of regions to rereplicate and amplify during S phase. Similar studies are also being conducted to better understand the role that chromatin control has in modulating DNA replication, transcription and genome integrity. The laboratory leverages a range of cytological, biochemical, genomic and epigenomic approaches to address DNA amplification control and DNA replication.
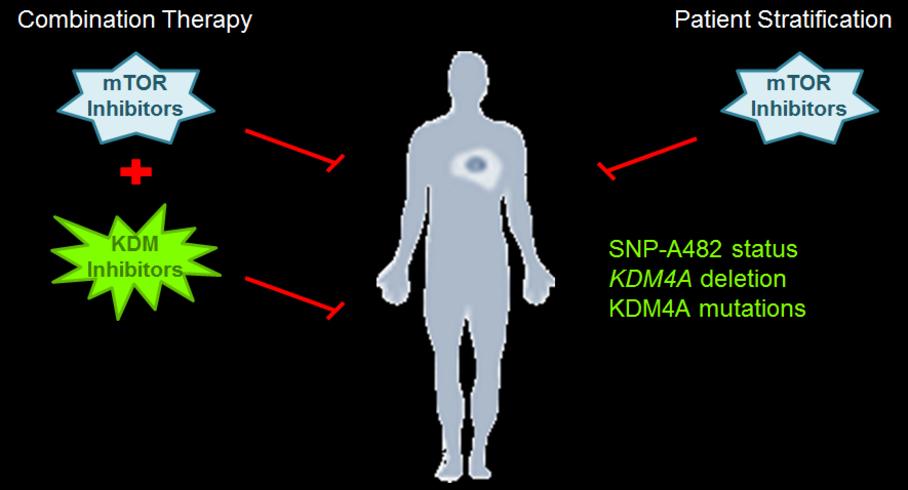
The Whetstine laboratory also addresses whether modulation of epigenetic factors provides novel therapeutic options across tumors (e.g., conventional chemotherapy and immune checkpoint therapy). His team is evaluating the expression levels, copy gains or losses, mutational and polymorphic repertoire in malignancies, as well as posttranslational regulatory mechanisms for epigenetic factors and how they relate to various tumor types. The Whetstine laboratory evaluates the impact that these events have on drug response and ability to stratify patients. These studies resulted in the identification of KDM4A associated copy gains as described above, as well as the identification of novel enzyme function for demethylases and association with chemotherapeutic response in lung cancer. Comparable studies are now being extended into additional chromatin modifying enzymes.
Key publications highlighting discoveries from the Whetstine Laboratory:
Black, J.C., Zhang, H., Kim, J., Getz, G., and Whetstine, J.R. (2016) Regulation of Transient Sitespecific
Copy Gain by MicroRNA. J. Biol. Chem. 291, 4862-4871
Black, J.C., Atabakhsh, E., Kim J., Biette K.B., Van Rechem C., Ladd B., Burrowes P.d., Donado C.,
Mattoo H., Kleinstiver B.P., Song B., Andriani G., Joung J.K., Iliopoulos O., Montagna C., Pillai S., Getz
G., Whetstine J.R. (2015) Hypoxia drives transient site-specific copy gan and drug-resistant gene
expression. Genes and Development 29, 1018-1031
Heinrichs, A. (2013) Chromatin affects Copy Gain. NSMB. 20, 1025.
Aranda, V. (2013) Unraveling chromatin checkpoints. Nat. Med. 19, 1102.
KDM4A Promotes Site-Specific Copy Number Gain (2013) CD-RW2013-162
Kiberstis, P.A. (2013) Gains in Cancer Epigenetics. Science. 341, 696.
Rickels, R., and Shilatifard, A. (2013) A Histone Modifier’s Ill-Gotten Copy Gains. Cell. 154, 477-479.
Black, J.C., Manning, A., Van Rechem, C., Kim, J., Ladd, B., Cho, J., Pineda, C.M., Murphy, N.,
Daniels, D.L., Montagna, S., Lewis, P.W., Glass, K., Allis, C.D., Dyson, N.J., Getz, G., Whetstine, J.R.
(2013) H3K9/36me3 Demethylase KDM4A Promotes Site-Specific Copy Gain and Re-replication of
Regions Amplified in Tumors. Cell. 154, 541-555. NIHMS525115
Lan, F., Bayliss, P. E.*, Rinn, J. L.*, Whetstine, J. R.*, Wang, J. K., Chen, S., Iwase, S., Alpatov, R.,
Issaeva, I., Canaani, E., Roberts, T. M., Chang, H. Y., and Shi, Y. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 2007; 449: 689-694. * Contributed Equal to this study.
Chen, Z.*, Zang, J.*, Whetstine, J. R., Hong, X., Davrazou, F., Kutateladze, T. G., Simpson, M., Mao,
Q., Pan, C-H., Dai, S., Hagman, J., Hansen, K., Shi, Y., and Zhang, G. (2006) Crystal Structure of the
Catalytic Core Domain of a Novel Histone Demethylase. Cell, 125, 691-702. * Contributed Equal to this study.
Whetstine, J. R., Nottke, A., Lan, F., Huarte, M., Smolikov, S., Chen, Z., Spooner, E., Li, E., Zhang, G.,
Colaiacovo, M., and Shi, Y. (2006) Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell, 125, 467-481.
Shi, Y., Lan, F., Matson, C., Mulligan, P., Whetstine, J. R., Cole, P. A., Casero, R. A., and Shi, Y.
(2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell, 119, 941-953. Collapse
Dr. Whetstine on PubMed
The Whetstine Laboratory at the Fox Chase Cancer Center is hiring highly ambitious and self-motivated Postdoctoral Fellows in Cancer Epigenetics. The laboratory uses state-of-the-art technologies and collaborations with leading scientist and clinicians to address the following areas: (1) the identification and characterization of epigenetic mechanisms driving extrachromosomal DNA amplification and chromosomal rearrangement, therapeutic resistance and tumor heterogeneity; (2) the impact of cancer-associated epigenetic factor variants (mutations and germline) on drug responses; and (3) the direct role of chromatin factors in regulating cell division, especially DNA replication. Some example publications addressing these areas of interest are: Black et al. (2010) Mol Cell; Black et al. (2013) Cell; Van Rechem et al. (2015) Cancer Discovery; Mishra et al. (2018) Cell; Clarke et al. (2020) Cancer Discov.; van Rechem et al. (2021) Cell Reports; Gray et al. (2023) Cell.
Learn more about the laboratory research in the Whetstine laboratory.
The Whetstine laboratory seeks candidates that published peer reviewed manuscripts and that have strong expertise in molecular biology, cancer biology, biochemistry, genomics, epigenomics and/or chromatin biology.
The Whetstine laboratory is a member of the Cancer Epigenetics Institute (CEI) at Fox Chase. The institute provides trainees unparalleled access to world-leaders in epigenetics as well as opportunities to interact with editors and industry leaders. CEI also has an annual symposium that brings leaders in the area of epigenetics from both academia and industry so that new advances are discussed. There are poster sessions and opportunities for travel awards at the annual symposium as well. Trainees also have ample opportunities to present their findings to colleagues in CEI and across FCCC. CEI aims to enhance each trainee’s opportunity for networking and exposure to leading science so they have an optimal opportunity to develop their career path.
As one of the four original cancer centers to receive comprehensive designation from the National Cancer Institute, Fox Chase Cancer Center has been at the forefront of cancer research for almost 90 years. We are home to excellent research facilities, top clinicians and scientists, and outstanding patient care. Our singular focus on cancer, which couples discovery science with state of the art clinical care and population health, remains the foundation of our work.
The scientist training programs at Fox Chase Cancer Center provide professional development opportunities in four core areas identified as crucial for successful careers in science, research, and health care including communication, leadership, teaching, and mentorship. Upon joining the program, graduate students and postdocs develop individual development plans to help guide their growth. Training throughout the year is supplemented with free professional development opportunities, including a robust ‘How To’ series, writing courses, networking, mentorship, and teaching opportunities, a trainee-led seminar series, a trainee-led annual Research Conference, and more. Postdocs at Fox Chase Cancer Center are supported by the Temple University Postdoc Association and the Office of Academic Affairs at Fox Chase, and are compensated with competitive pay and benefits.
In addition to the robust training program, scientists at Fox Chase Cancer Center benefit from being part of the rich scientific and biotech environment in the Philadelphia region. Many of our former trainees are now employees (and contacts) at nearby institutions and companies, including The Wistar Institute, Merk, GSK, AACR, and numerous others.
Email a CV, cover letter that states research interest(s) and goals, and the name of at least three references to [email protected].
This Fox Chase professor participates in the Undergraduate Summer Research Fellowship.
Learn more about Research Volunteering.