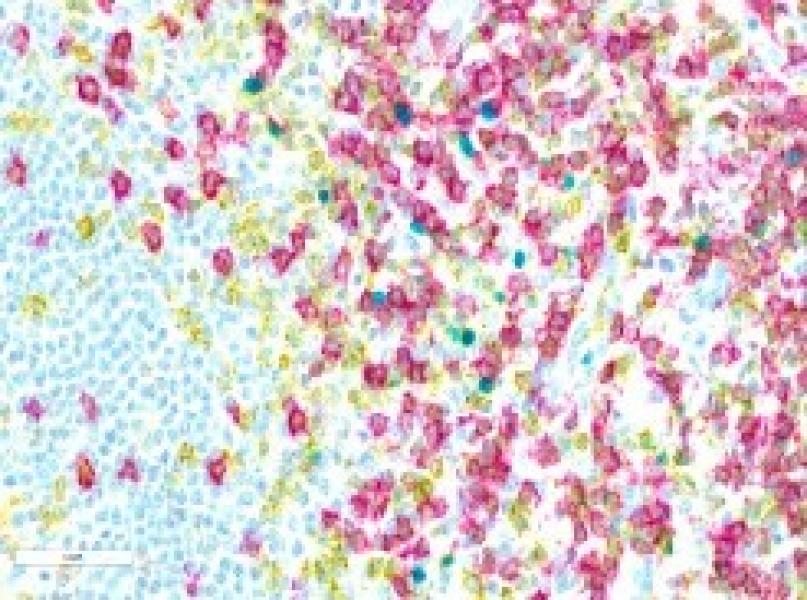
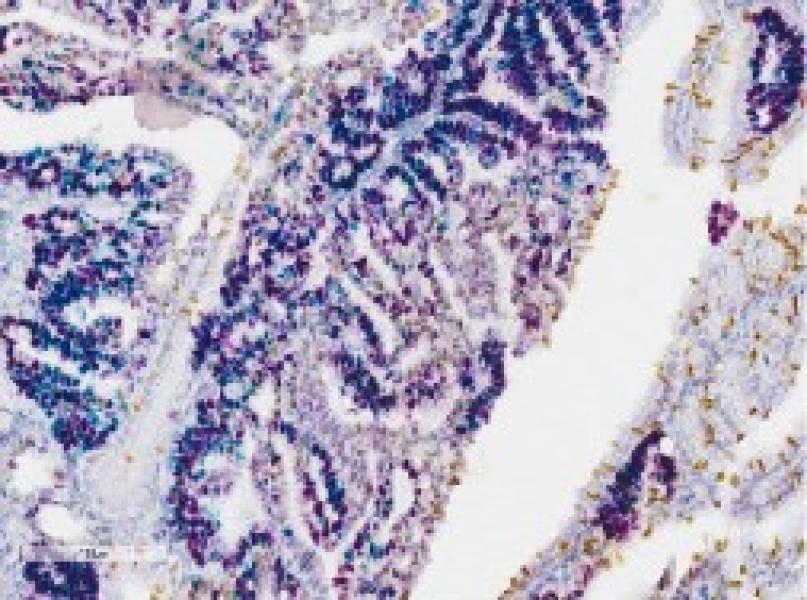
This facility derives partial funding from the FCCC Cancer Center Support Grant (CCSG) from the National Cancer Institute.
|
Director |
Kathy Q. Cai, MD, PhD [email protected] Office: W232 215-214-3208 |
| Histopathology Manager | Jirong (Jenny) Zhang [email protected] Office: W232 215-728-2464 |
| Histopathology Technical Specialist: (ASCP certified histotechnologist) | Nina Ibeme [email protected] Office: W232 215-728-2464 |
| Research Associate | Li Wang [email protected] Office: W230 215-728-2464 |
| Shipping Address | Histopathology Facility 333 Cottman Avenue, W232 Philadelphia, PA 19111 |
| Consultant | Andres Klein-Szanto, M.D., Ph.D. [email protected] 215-728-3154 Office: W230 |
| Pricing & Scheduling | Contact Jirong (Jenny) Zhang for pricing and scheduling |
The Histopathology Facility (HF) provides Cancer Center members with technical processing of cells and tissues, gene expression in situ protein immunolocalization, pathology interpretation of microscopy findings, including target distribution and other metrics focused on particular cell/tissue compartments. We help establish and characterize animal models of human cancer and related diseases, as well as analysis of human tissue samples, including from clinical trials. Most projects involve animal models that require complex histological, immunohistochemical and/or cytological processing to determine morphological alterations, and to localize and quantify the expression of cancer-relevant gene products in tissue sections, including murine and human samples.
The major objective of the animal/human research services of the Histopathology and Facility (HF) is to facilitate research conducted by NCI/NIH-funded Cancer Center members by providing them with technical processing of cells and tissues as well as issuing reports of histopathological and immunohistochemical (IHC) /image analysis findings. Primary activities include:
- Preparation of tissues for histopathological or cytological evaluation, i.e., embedding cells and tissues in paraffin, preparation of tissue sections, and staining. Preparation of frozen sections, histochemistry, IHC, including multi-chromatic IHC, and in situ hybridization. Provision of slide scanning and IHC image analysis as well as Laser Capture Microdissection (LCM).
- Provision of interpretation and consultation on procedures, results and histopathological evaluations. The Facility also provides documentation on techniques and reports results. In addition, we document results using digital photography and work with Cancer Center members to devise custom-made approaches for projects as needed.