Related Articles
00 / 00

This Fox Chase professor participates in the Undergraduate Summer Research Fellowship.
Learn more about Research Volunteering.
Marvin & Concetta Greenberg Chair in Pancreatic Cancer Research
ACS Wilmott Family Professor of Pancreatic Cancer
Co-Leader, Cancer Signaling and Microenvironment Program
Director, Spatial Immuno-Proteomic Initiative (Highplex Spatial Immunofluorescence)
The Cukierman Laboratory is focused on understanding how desmoplasia, the dense mesenchymal microenvironment surrounding solid epithelial tumors, affects tumor progression. The team uses a unique 3D culturing system that mimics the in vivo mesenchymal stroma to study the functional roles of desmoplasia in tumorigenic behaviors. They have developed Harmonic Output of Stromal Traits (HOST), a novel method for identifying tumor microenvironment (TME) cells and HOST-Factor, a numerical metric that quantifies their functional states. These tools allow the laboratory to assess the relative contribution of the main functional unit composed of cancer-associated fibroblasts (CAFs) and CAF-generated extracellular matrix (ECM), as well as the units' interaction with other TME cells (e.g., immune cells, nerves, cancer cells and more) to impart tumor-supportive or tumor-suppressive functions. They are also investigating whether HOST-Factor values can serve as TME-based prognostic indicators for patient outcomes and predict or inform on drug efficacy.
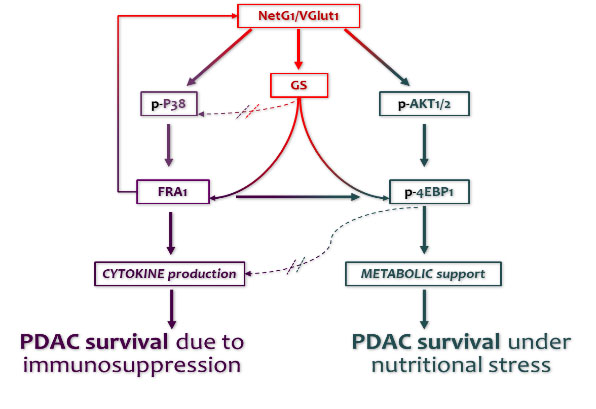
Researchers at Fox Chase Cancer Center have identified a new biomarker and potential novel treatment target for pancreatic ductal adenocarcinoma (PDAC) that could help starve cancer cells and allow the immune system to attack the tumor. Read the full article here.









Former Personnel
Ashish Abraham
Jennifer Alexander
Michael Amatangelo
Daniel Bassi
Dorothy Beacham
Leila Borghaei
Kimberly Brown
Miranda Burdiel-Herencia
Remedios Castelló-Cros
Lauren Cheifitz
Gil Cukierman
Jonte Desire
Xiaoshen Dong
Stephanie Elbe
James Flaherty
Ralph Francescone
Sarah Goldston
Vivekanad Gupta
Kelci R. Holman
David Khan
Melissa Lech
Raj Madhani
Ruchi Malik
Emily Malloy
Renato Mattos
Karen Melo
Charline Ogier
Christopher Price
Cassidy Poon
Roderick Quiros
Kristopher Raghavan
Poornima Rao
Mastan Rao Chintalapudi
Margot Ruan
Rebecca Roth
Dustin Rollins
Brad Rybinski
Neelima Shah
Jeffery Simons
Jerry So
Julie Sosa
Matthildi Valianou
Debora B Vendramini Costa
Fernanda Villamar
Olga Villamar
Corinne Watson
Gary Wilk
Galia Wilk
Stephanie Wirtshafter
Daniel Zinshteyn
Alexander Zenin
Desmoplastic Tumor Microenvironment (TME)
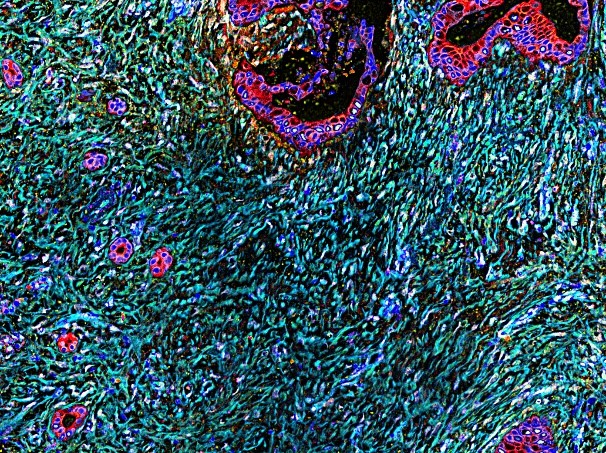
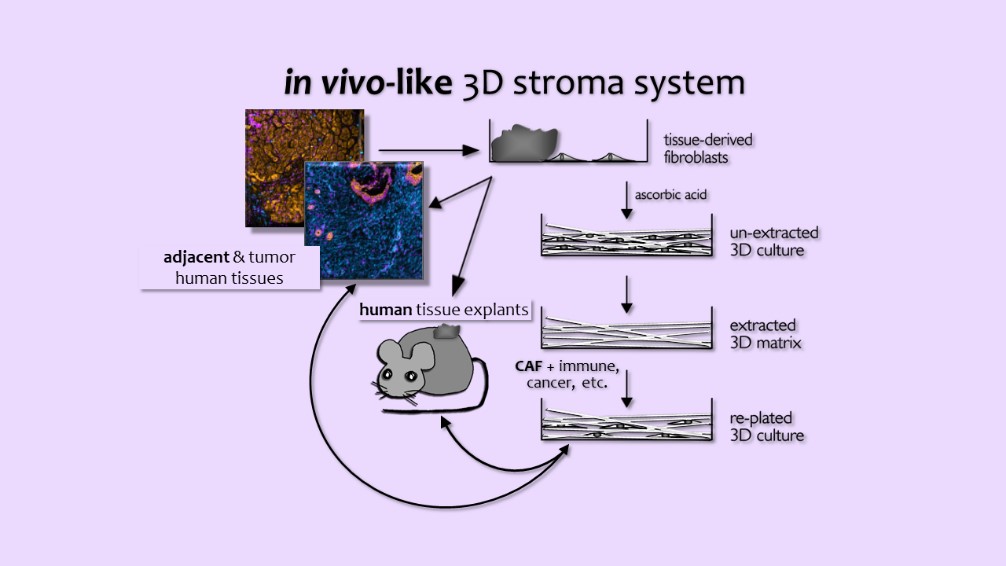
The Cukierman Laboratory is dedicated to understanding the complex interplay between desmoplasia, the dense mesenchymal microenvironment of solid tumors, and tumor progression. Utilizing a unique 3D culturing system that mimics the in vivo mesenchymal stroma, the laboratory investigates the functional roles of desmoplasia in tumorigenic behaviors, including invasive spread, metabolic support, and immune suppression.
Leveraging a multifaceted approach that integrates cell biological and bioengineering techniques, the Cukierman team has developed Harmonic Output of Stromal Traits (HOST), a novel method for identifying tumor microenvironment (TME) cells and HOST-Factor, a numerical metric that quantifies their functional states. These tools enable the laboratory to assess the relative contribution of the main functional unit composed of cancer-associated fibroblasts (CAFs) and CAF-generated extracellular matrix (ECM), as well as the units’ interaction with other TME cells to impart tumor-supportive or tumor-suppressive functions.
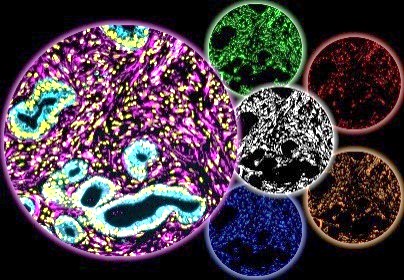
Ongoing research within the Cukierman Laboratory seeks to uncover key mechanisms of CAF/ECM unit functions, identify new stromal targets, and develop stroma targeting modalities. Additionally, the laboratory is investigating whether HOST-Factor values can serve as TME-based prognostic indicators for patient outcomes and predict or inform on drug efficacy. Moreover, the team is exploring the potential influence of macro-environmental factors, such as residing in low socioeconomic status (nSES) neighborhoods, in exacerbating the pro-cancer TME function.
In summary, the Cukierman Laboratory is at the forefront of desmoplasia research, employing a unique 3D culturing system, novel analytical tools, and a multifaceted approach to unravel the complexities of this critical TME component. The laboratory's findings hold promise for the development of novel therapeutic strategies that target the desmoplasia-tumor interactions, ultimately improving patient outcomes for solid epithelial cancers.