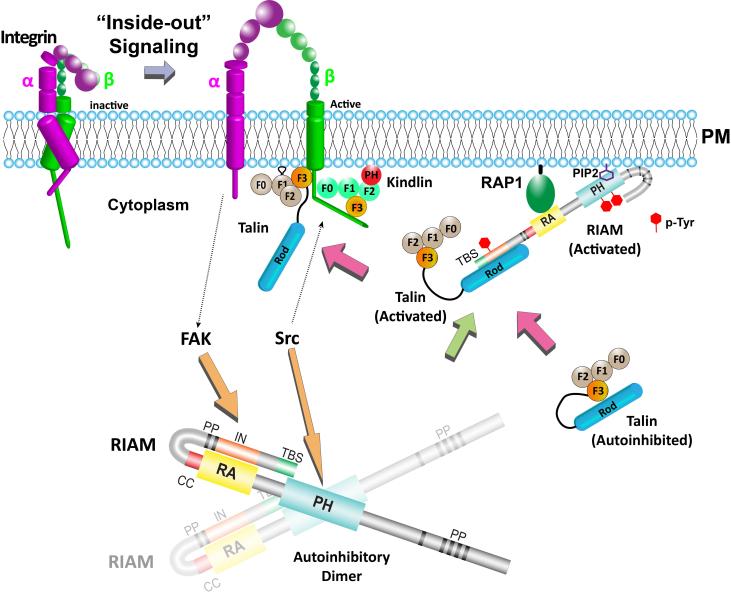
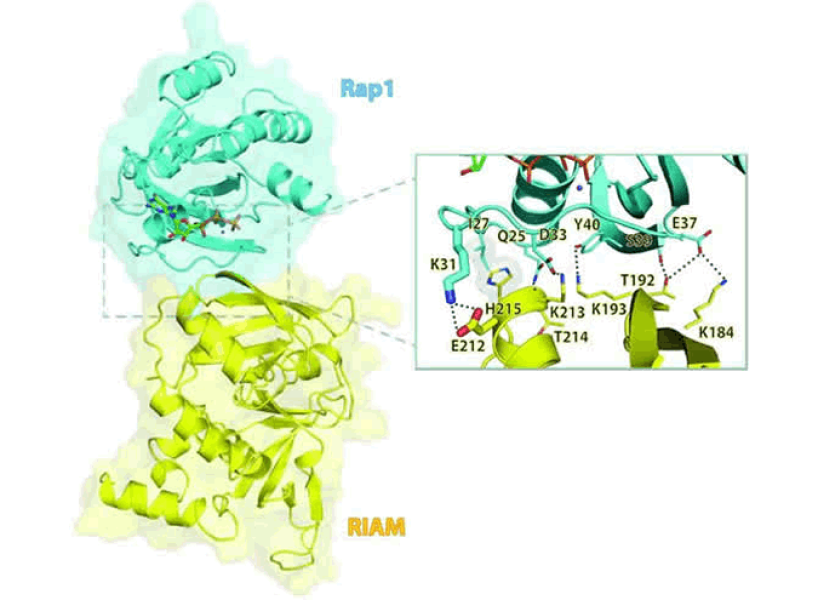
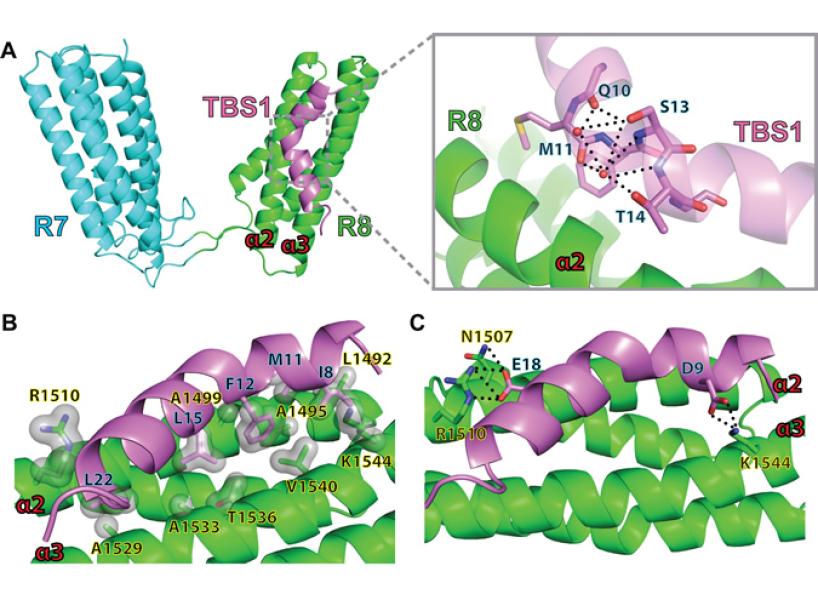
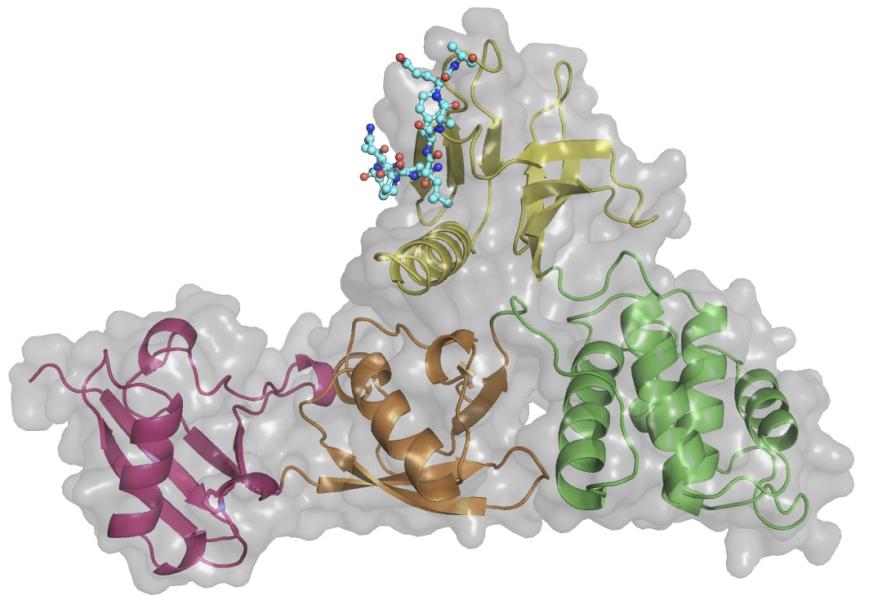
This Fox Chase professor participates in the Undergraduate Summer Research Fellowship.
Learn more about Research Volunteering.
Weather Alert: Following the winter storm, all Temple Health hospitals, campuses and clinical locations remain open. Patients will be contacted directly if their visit is affected. Please check TempleHealth.org or FoxChase.org for updates and monitor myTempleHealth for changes to scheduled appointments.