Breadcrumb
- Home
- Mitchell Fane
Mitchell E Fane, PhD

Educational Background
- B.S., Biomedical Science (Honors Class I), University of Queensland, Brisbane, Australia
- Doctor of Philosophy (University of Queensland, School of Biomedical Sciences)
- Research project: Investigating the role of the MITF-BRN2 expression axis in melanoma. Advisors: Dr Aaron Smith, Dr Richard Sturm
Memberships
- International Pigment Cell Conference
- Society of Melanoma Research
- American Association of Cancer Research
Honors & Awards
- Larry Grossman Award for Excellence in Postdoctoral Research, Johns Hopkins School of Public Health (2021)
- Best abstract / Travel award for the Society for Melanoma Research, Virtual congress (2021)
- Travel award and proffered talk at Annual AACR Scientific Meeting San Diego, California, United States of America (2020)
- Proffered talk at Society of Melanoma Research Conference Utah, United States of America, 2019
- Travel award and invited speaker at the GSA Annual Scientific Meeting Austin, Texas, United States of America (2019)
- Wistar Institute International Travel Award, Summer, Manchester, United Kingdom, 2018
- International travel award and proffered talk for AACR Cancer Dormancy and Residual Disease Conference, Montreal, Quebec, Canada, (2018)
- International travel award for the Society for Melanoma Research, Boston, Massachusetts, United States of America, (2016)
- SBMS international travel award UQ Brisbane, Australia (2016)
- Best oral Presentation UQ SBMS Conference, Brisbane, Australia (2015)
- Invited talk and travel award to the International Pigment Cell Conference, Singapore, (2014)
- Best poster Presentation UQ SBMS Conference, Brisbane, Australia (2014)
- APA PhD Scholarship UQ, Brisbane, Australia, (2013-2017)
- SBMS Honors Bursary ($5000) UQ, Brisbane, Australia, (2012)
- UQ Research Summer Scholarship ($2400), Brisbane, Australia, (2012, 2013)
- Honors Bursary from the Asthma foundation ($5000), Brisbane, Australia, (2011)
- 4 Dean Commendations UQ, Brisbane, Australia, (2009-2011)
Research Interests
Fane Lab – The role of aging on metastatic niche changes and cancer dormancy
- Deciphering the intrinsic mechanisms of melanoma dormancy and reactivation in metastatic niches of young and aged patient and pre-clinical animal models.
- Investigating fibroblast, ECM and immune-mediated changes associated with aging in healthy and cancer containing metastatic niches.
- Defining differences in secretion of soluble proteins from dormant vs reactivated metastatic cancer cells and their role in cancer progression.
- Uncovering the role of aging in promoting resistance to targeted and immune checkpoint therapies across differing metastatic niches.
Lab Overview
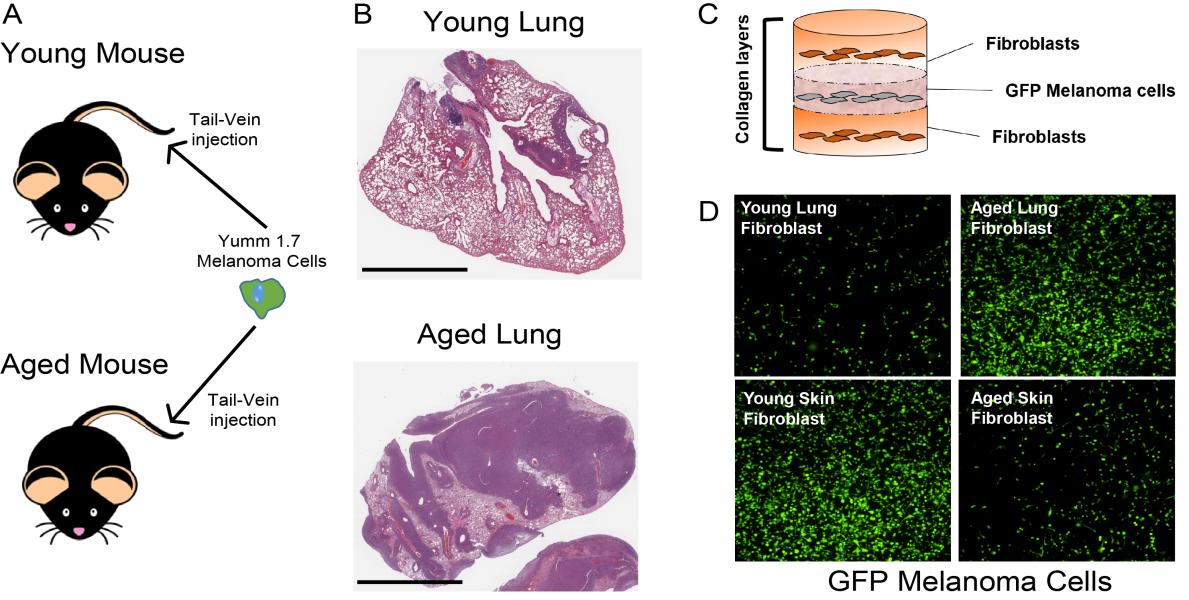
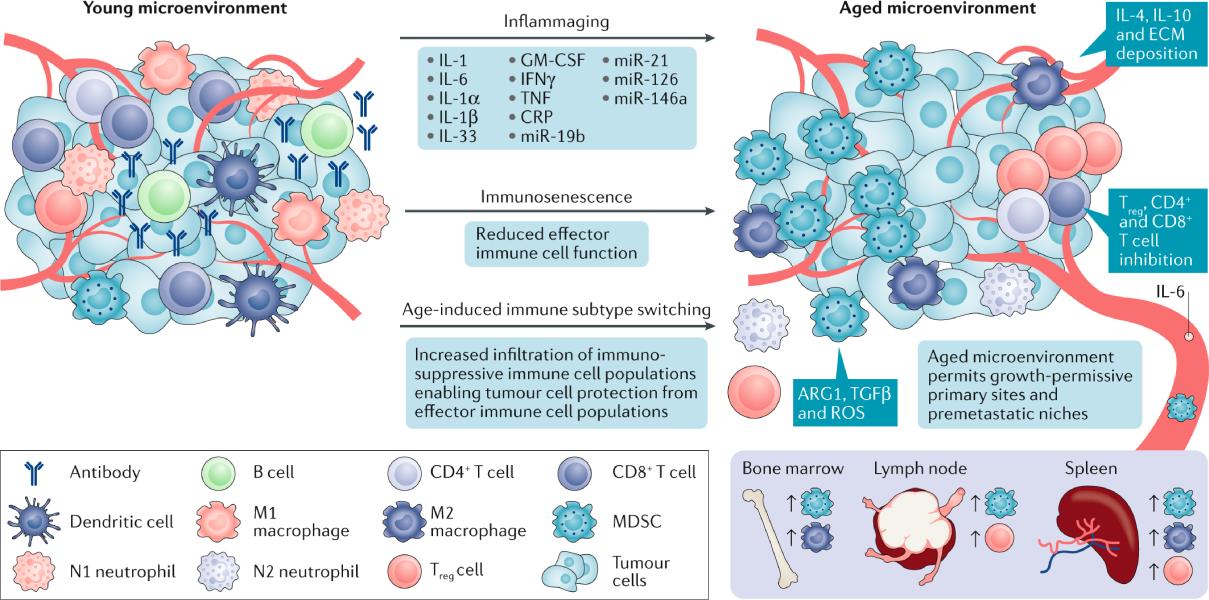
The research program within the Fane lab focuses on trying to understand the molecular mechanisms involved in the aging pre-metastatic niches and the role of these aged microenvironments in promoting cancer cell dormancy, metastatic reactivation and response to therapy.
We are interested in uncovering the evolution and intrinsic reprograming of fibroblasts, immune cells, endothelial cells and ECM remodeling associated with aging in the pre-metastatic niche of healthy and cancerous in-vitro and in-vivo models of metastasis, with validation using patient samples.
Our goal is to uncover novel signaling pathways associated with dormant and reactivated cancerous disease across various aging metastatic cancer models, with the hope of uncovering clinical markers for diagnosis of dormant disease and therapies for targeting these highly resistant and eventually aggressive cancer populations.
We also place a large emphasis on investigating the use of aged models across cancer. Many cancers show dramatic increases in incidence and mortality with respect to age, yet most pre-clinical animal models are done in younger mice or do not take account of age within patient samples. By deciphering the key differences between young and aged microenvironments in healthy and diseased models, we hope to create a paradigm shift in the field where age appropriate models are considered for various cancers for better clinical translation.
Selected Publications
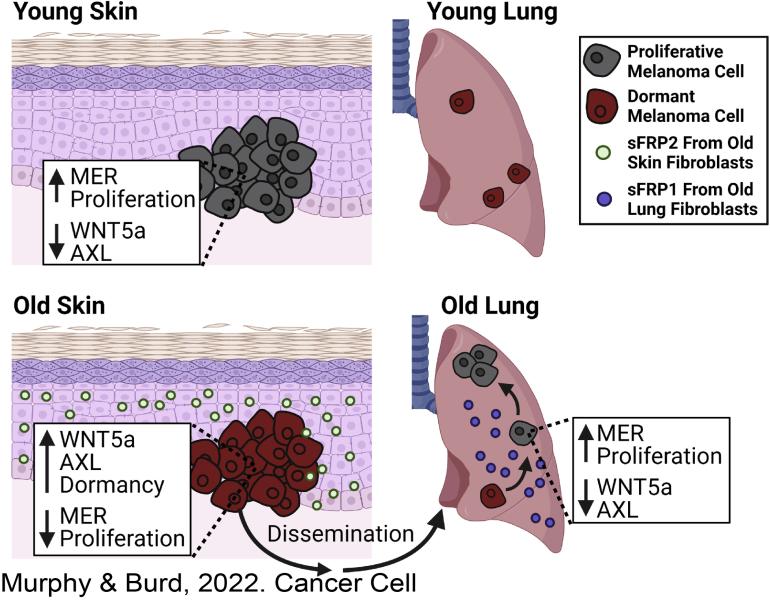
Fane, M.E ,Yash Chhabra, Gretchen M. Alicea, Devon A. Maranto, Stephen M. Douglass, Marie R. Webster, Vito W. Rebecca, Gloria E. Marino, Filipe Almeida, Brett L. Ecker, Daniel J. Zabransky, Laura Hüser, Thomas Beer, Hsin-Yao Tang, Andrew Kossenkov, Meenhard Herlyn, David W. Speicher, Wei Xu, Xiaowei Xu, Elizabeth M. Jaffee, Julio A. Aguirre-Ghiso, & Ashani T. Weeraratna. Stromal Changes in The Aged Lung Induce an Emergence From Melanoma Dormancy. Nature. 2022 June 01. https://doi.org/10.1038/s41586-022-04774-2.
Fane ME, Chhabra Y, Spoerri L, Simmons JL, Ludwig R, Bonvin E, Goding CR, Sturm RA, Boyle GM, Haass NK, Piper M, Smith AG. Reciprocal Regulation of BRN2 and NOTCH1/2 Signaling Synergistically Drives Melanoma Cell Migration and Invasion. J Invest Dermatol. 2021 Dec 24;. doi: 10.1016/j.jid.2020.12.039. [Epub ahead of print] PubMed PMID: 34958806.
Fane ME, Ecker BL, Kaur A, Marino GE, Alicea GM, Douglass SM, Chhabra Y, Webster MR, Marshall A, Colling R, Espinosa O, Coupe N, Maroo N, Campo L, Middleton MR, Corrie P, Xu X, Karakousis GC, Weeraratna AT. sFRP2 Supersedes VEGF as an Age-related Driver of Angiogenesis in Melanoma, Affecting Response to Anti-VEGF Therapy in Older Patients. Clin Cancer Res. 2020 Nov 1;26(21):5709-5719. doi: 10.1158/1078-0432.CCR-20-0446. PubMed PMID: 33097493; PubMed Central PMCID: PMC7642114.
*Fane M, Weeraratna AT. How the ageing microenvironment influences tumour progression. Nat Rev Cancer. 2020 Feb;20(2):89-106. doi: 10.1038/s41568-019-0222-9. Epub 2019 Dec 13. Review. PubMed PMID: 31836838; PubMed Central PMCID: PMC7377404.
Webster MR, Fane ME, Alicea GM, Basu S, Kossenkov AV, Marino GE, Douglass SM, Kaur A, Ecker BL, Gnanapradeepan K, Ndoye A, Kugel C, Valiga A, Palmer J, Liu Q, Xu X, Morris J, Yin X, Wu H, Xu W, Zheng C, Karakousis GC, Amaravadi RK, Mitchell TC, Almeida FV, Xiao M, Rebecca VW, Wang YJ, Schuchter LM, Herlyn M, Murphy ME, Weeraratna AT. Paradoxical Role for Wild-Type p53 in Driving Therapy Resistance in Melanoma. Mol Cell. 2020 Feb 6;77(3):681. doi: 10.1016/j.molcel.2020.01.005. PubMed PMID: 32032511; PubMed Central PMCID: PMC7441646.
Douglass SM, Fane ME, Sanseviero E, Ecker BL, Kugel CH 3rd, Behera R, Kumar V, Tcyganov EN, Yin X, Liu Q, Chhabra Y, Alicea GM, Kuruvilla R, Gabrilovich DI, Weeraratna AT. Myeloid-Derived Suppressor Cells Are a Major Source of Wnt5A in the Melanoma Microenvironment and Depend on Wnt5A for Full Suppressive Activity. Cancer Res. 2021 Feb 1;81(3):658-670. doi: 10.1158/0008-5472.CAN-20-1238. Epub 2020 Dec 1. PubMed PMID: 33262126; PubMed Central PMCID: PMC8330365.
Alicea GM, Rebecca VW, Goldman AR, Fane ME, Douglass SM, Behera R, Webster MR, Kugel CH 3rd, Ecker BL, Caino MC, Kossenkov AV, Tang HY, Frederick DT, Flaherty KT, Xu X, Liu Q, Gabrilovich DI, Herlyn M, Blair IA, Schug ZT, Speicher DW, Weeraratna AT. Changes in Aged Fibroblast Lipid Metabolism Induce Age-Dependent Melanoma Cell Resistance to Targeted Therapy via the Fatty Acid Transporter FATP2. Cancer Discov. 2020 Sep;10(9):1282-1295. doi: 10.1158/2159-8290.CD-20-0329. Epub 2020 Jun 4. PubMed PMID: 32499221; PubMed Central PMCID: PMC7483379.
Torre EA, Arai E, Bayatpour S, Jiang CL, Beck LE, Emert BL, Shaffer SM, Mellis IA, Fane ME, Alicea GM, Budinich KA, Weeraratna AT, Shi J, Raj A. Genetic screening for single-cell variability modulators driving therapy resistance. Nat Genet. 2021 Jan;53(1):76-85. doi: 10.1038/s41588-020-00749-z. Epub 2021 Jan 4. PubMed PMID: 33398196; PubMed Central PMCID: PMC7796998.
Dardani I, Emert BL, Goyal Y, Jiang CL, Kaur A, Lee J, Rouhanifard SH, Alicea GM, Fane ME, Xiao M, Herlyn M, Weeraratna AT, Raj A. ClampFISH 2.0 enables rapid, scalable amplified RNA detection in situ. Nature Methods. 2022;19(11):1403-10. doi: 10.1038/s41592-022-01653-6.
Fane ME, Chhabra Y, Hollingsworth DEJ, Simmons JL, Spoerri L, Oh TG, Chauhan J, Chin T, Harris L, Harvey TJ, Muscat GEO, Goding CR, Sturm RA, Haass NK, Boyle GM, Piper M, Smith AG. NFIB Mediates BRN2 Driven Melanoma Cell Migration and Invasion Through Regulation of EZH2 and MITF. EBioMedicine. 2017 Feb;16:63-75. doi: 10.1016/j.ebiom.2017.01.013. Epub 2017 Jan 16. PubMed PMID: 28119061; PubMed Central PMCID: PMC5474438.
Kugel CH 3rd, Douglass SM, Webster MR, Kaur A, Liu Q, Yin X, Weiss SA, Darvishian F, Al-Rohil RN, Ndoye A, Behera R, Alicea GM, Ecker BL, Fane M, Allegrezza MJ, Svoronos N, Kumar V, Wang DY, Somasundaram R, Hu-Lieskovan S, Ozgun A, Herlyn M, Conejo-Garcia JR, Gabrilovich D, Stone EL, Nowicki TS, Sosman J, Rai R, Carlino MS, Long GV, Marais R, Ribas A, Eroglu Z, Davies MA, Schilling B, Schadendorf D, Xu W, Amaravadi RK, Menzies AM, McQuade JL, Johnson DB, Osman I, Weeraratna AT. Age Correlates with Response to Anti-PD1, Reflecting Age-Related Differences in Intratumoral Effector and Regulatory T-Cell Populations. Clin Cancer Res. 2018 Nov 1;24(21):5347-5356. doi: 10.1158/1078-0432.CCR-18-1116. Epub 2018 Jun 13. PubMed PMID: 29898988; PubMed Central PMCID: PMC6324578.
Ecker BL, Kaur A, Douglass SM, Webster MR, Almeida FV, Marino GE, Sinnamon AJ, Neuwirth MG, Alicea GM, Ndoye A, Fane M, Xu X, Sim MS, Deutsch GB, Faries MB, Karakousis GC, Weeraratna AT. Age-Related Changes in HAPLN1 Increase Lymphatic Permeability and Affect Routes of Melanoma Metastasis. Cancer Discov. 2019 Jan;9(1):82-95. doi: 10.1158/2159-8290.CD-18-0168. Epub 2018 Oct 2. PubMed PMID: 30279172; PubMed Central PMCID: PMC6328344.
Kaur A, Ecker BL, Douglass SM, Kugel CH 3rd, Webster MR, Almeida FV, Somasundaram R, Hayden J, Ban E, Ahmadzadeh H, Franco-Barraza J, Shah N, Mellis IA, Keeney F, Kossenkov A, Tang HY, Yin X, Liu Q, Xu X, Fane M, Brafford P, Herlyn M, Speicher DW, Wargo JA, Tetzlaff MT, Haydu LE, Raj A, Shenoy V, Cukierman E, Weeraratna AT. Remodeling of the Collagen Matrix in Aging Skin Promotes Melanoma Metastasis and Affects Immune Cell Motility. Cancer Discov. 2019 Jan;9(1):64-81. doi: 10.1158/2159-8290.CD-18-0193. Epub 2018 Oct 2. PubMed PMID: 30279173; PubMed Central PMCID: PMC6328333.
Fane ME, Chhabra Y, Smith AG, Sturm RA. BRN2, a POUerful driver of melanoma phenotype switching and metastasis. Pigment Cell Melanoma Res. 2019 Jan;32(1):9-24. doi: 10.1111/pcmr.12710. Epub 2018 Jun 5. Review. PubMed PMID: 29781575
Open Positions
About the Position
A postdoctoral position is available to study the molecular mechanisms involved in how aging can induce reactivation from metastatic dormancy. The individual will be involved in using genetic animal models coupled with transcriptomic, proteomic and molecular biology techniques to investigate changes in melanoma cell signaling within an aged metastatic niche, along with changes in immune cell filtration, ECM deposition and fibroblast protein secretion. The long-term goal is to uncover clinical and prognostic markers of dormancy and reactivation within a metastatic niche and the potential to therapeutically target each melanoma cell population and/or stromal components in the tumor microenviroment, with the aim of creating more durable therapies for metastatic melanoma or increasing efficacy of current immunotherapies.
The primary techniques will involve western blot, immunohistochemistry, flow-cytometry, tissue culture, and mouse work. The individual will have the opportunity to learn and perform innovative techniques including CyTOF, Imaging Mass Cytometry, spatial transcriptomics and single-cell RNA-Seq. They will also gain extensive training in mouse modelling and lentiviral production and transduction. Interested applicants should have an interest in cancer cell signaling, aging tumor microevironment, dormancy and immune responses to cancer and ideally familiarity with mouse modelling and cell culture would be desirable. The individual will also require a doctoral degree.
About the Training Environment
As one of the four original cancer centers to receive comprehensive designation from the National Cancer Institute, Fox Chase Cancer Center has been at the forefront of cancer research for almost 90 years. We are home to excellent research facilities, top clinicians and scientists, and outstanding patient care. Our singular focus on cancer, which couples discovery science with state of the art clinical care and population health, remains the foundation of our work.
The scientist training programs at Fox Chase Cancer Center provide professional development opportunities in four core areas identified as crucial for successful careers in science, research, and health care including communication, leadership, teaching, and mentorship. Upon joining the program, graduate students and postdocs develop individual development plans to help guide their growth. Training throughout the year is supplemented with free professional development opportunities, including a robust ‘How To’ series, writing courses, networking, mentorship, and teaching opportunities, a trainee-led seminar series, a trainee-led annual Research Conference, and more. Postdocs at Fox Chase Cancer Center are supported by the Temple University Postdoc Association and the Office of Academic Affairs at Fox Chase, and are compensated with competitive pay and benefits.
In addition to the robust training program, scientists at Fox Chase Cancer Center benefit from being part of the rich scientific and biotech environment in the Philadelphia region. Many of our former trainees are now employees (and contacts) at nearby institutions and companies, including The Wistar Institute, Merck, GSK, AACR, and numerous others.
To Apply
Please email a CV, a cover letter and a brief description of your research interest, goals and the name and contact details of at least two references to Dr. Mitchell Fane ([email protected]).