This Fox Chase professor participates in the Undergraduate Summer Research Fellowship.
Learn more about Research Volunteering.

This Fox Chase professor participates in the Undergraduate Summer Research Fellowship.
Learn more about Research Volunteering.


Epigenetic mechanisms of gene expression and regulation
Development and functioning of higher organisms critically depends on properly regulated gene expression. Regulation of gene expression occurs primarily at the initial step (transcription) and involves DNA sequences, protein factors and dynamic changes in structure of DNA-protein complexes (chromatin). The major research goal in my laboratory is to understand the molecular mechanisms and regulation of the vital process of eukaryotic transcription in chromatin, and the role of the factors involved in cancer development and human aging (e.g. hFACT and hPARP1) in this process. This goal will be achieved using a combination of molecular genetics, genomics, biochemical, single-particle, structural and computational modeling approaches.
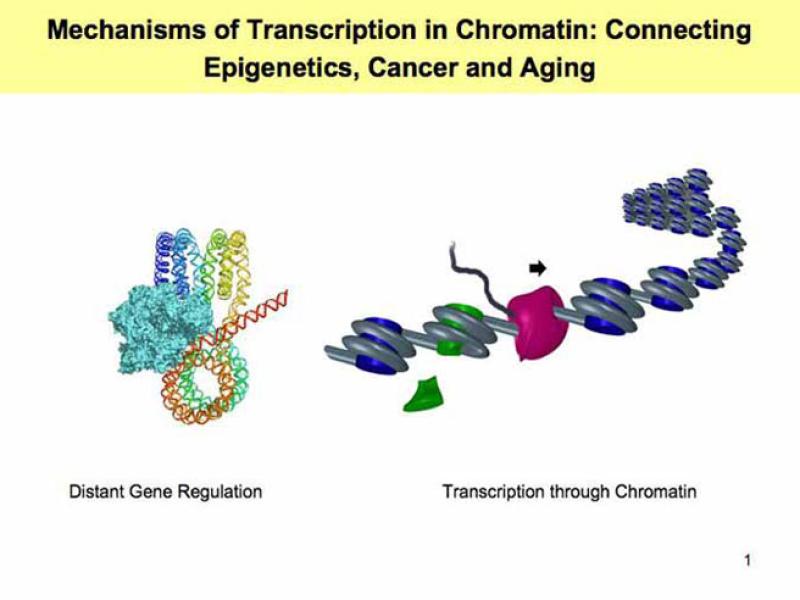
Development and functioning of higher organisms critically depends on properly regulated gene expression. Regulation of gene expression occurs primarily at the initial step (transcription) and involves DNA sequences, protein factors and dynamic changes in structure of DNA-protein complexes (chromatin). The major research goal in my laboratory is to understand the molecular mechanisms and regulation of the vital process of eukaryotic transcription in chromatin, and the role of the factors involved in cancer development and human aging (e.g. hFACT and hPARP1) in this process. This goal will be achieved using a combination of molecular genetics, genomics, biochemical, single-particle, structural and computational modeling approaches. Currently our efforts are focused in the following primary directions: 1. Mechanisms of histone chaperones FACT and PARP1 action during transcription and its regulation. 2. Mechanisms of interaction of sequence-specific regulator proteins (e.g. p53) with DNA organized into chromatin. 3. The mechanisms and regulation of transcript elongation through chromatin by RNA polymerase II, and 4. The mechanisms of gene regulation over a distance (by enhancers & insulators).
FACT and PARP1 are histone chaperones that can increase chromatin accessibility to various proteins during initiation and elongation of transcription. Importantly, both hFACT & hPARP1 are involved in carcinogenesis and are important targets for anti-cancer drugs. Recently we have studied the mechanisms of FACT- and PARP1-dependent chromatin reorganization. Our research focuses on the following questions: What changes in chromatin structure are involved in the reorganization? How do various anti-cancer drugs affect this process? What are the mechanisms of trapping of the histone chaperones in chromatin?
Tumor suppressor p53 is a sequence-specific DNA-binding protein, possibly a pioneering factor involved in gene regulation. It has an apparent high affinity to the response elements regulating cell cycle arrest genes (CCA-sites) and a lower affinity to the sites associated with apoptosis (Apo-sites) in vivo. We are trying to understand the mechanisms of interaction of p53 with its binding sites in chromatin. The questions are: Can the differences in affinity be explained by the chromatin context of the binding sites? What changes in chromatin structure are involved in p53-DNA interaction?
In early 2000 we have established a “minimal” experimental system that maintains transcription through various defined mono- and polynucleosomes by RNA polymerase II. This system faithfully recapitulates numerous features of transcribed chromatin described in vivo and allows their molecular analysis in vitro. Using this system, we have discovered a novel mechanism involving survival of core histones and their modifications without even transient histone dissociation from DNA. These features of the mechanism suggest that it is likely used for maintenance of chromatin structure and the “histone code”. The most recent focus of our studies is on the mechanism of action of histone chaperones FACT & PARP1 during Pol II transcription.
Distant regulation of gene transcription initiation is mediated by direct interaction between proteins bound at communicating DNA elements and involves looping of intervening DNA/chromatin regions. Our research focuses on the following critical questions: What features of DNA/chromatin template allow efficient communication over a distance? What are distinct features of the regulatory elements that are capable of action over a distance? We have adopted relatively simple, highly purified and efficient experimental systems for quantitative analysis of enhancer action over a distance in vitro. This simple experimental system allowed us to identify elements of chromatin structure and protein factors involved in distant gene regulation.
Valieva ME, Gerasimova NS, Kudryashova KS, Kozlova AL, Kirpichnikov MP, Hu Q, Botuyan MV, Mer G, Feofanov AV, Studitsky VM. Stabilization of Nucleosomes by Histone Tails and by FACT Revealed by spFRET Microscopy. Cancers (Basel). 2017;9(1):pii: E3. Epub 2017/01/10. doi: 10.3390/cancers9010003. PubMed PMID: 28067802; PMCID: PMC5295774.
Sultanov DC, Gerasimova NS, Kudryashova KS, Maluchenko NV, Kotova EY, Langelier MF, Pascal JM, Kirpichnikov MP, Feofanov AV, Studitsky VM. Unfolding of core nucleosomes by PARP-1 revealed by spFRET microscopy. AIMS Genet. 2017;4(1):21-31. Epub 2017/08/15. doi: 10.3934/genet.2017.1.21. PubMed PMID: 28804761; PMCID: PMC5552189.
Nizovtseva EV, Todolli S, Olson WK, Studitsky VM. Towards quantitative analysis of gene regulation by enhancers. Epigenomics. 2017;9(9):1219-31. Epub 2017/08/12. doi: 10.2217/epi-2017-0061. PubMed PMID: 28799793; PMCID: PMC5585842.
Nizovtseva EV, Clauvelin N, Todolli S, Polikanov YS, Kulaeva OI, Wengrzynek S, Olson WK, Studitsky VM. Nucleosome-free DNA regions differentially affect distant communication in chromatin. Nucleic Acids Res. 2017;45(6):3059-67. Epub 2016/12/13. doi: 10.1093/nar/gkw1240. PubMed PMID: 27940560; PMCID: PMC5389534.
Chang HW, Studitsky VM. Chromatin replication: TRANSmitting the histone code. Journal of nature and science. 2017;3(2):pii: e322. Epub 2017/04/11. PubMed PMID: 28393112; PMCID: PMC5384335.
Valieva ME, Armeev GA, Kudryashova KS, Gerasimova NS, Shaytan AK, Kulaeva OI, McCullough LL, Formosa T, Georgiev PG, Kirpichnikov MP, Studitsky VM, Feofanov AV. Large-scale ATP-independent nucleosome unfolding by a histone chaperone. Nat Struct Mol Biol. 2016;23(12):1111-6. Epub 2016/11/08. doi: 10.1038/nsmb.3321. PubMed PMID: 27820806; PMCID: PMC5518926.
Studitsky VM, Nizovtseva EV, Shaytan AK, Luse DS. Nucleosomal Barrier to Transcription: Structural Determinants and Changes in Chromatin Structure. Biochemistry & molecular biology journal. 2016;2(2). Epub 2016/10/19. doi: 10.21767/2471-8084.100017. PubMed PMID: 27754494; PMCID: PMC5041593.
Gerasimova NS, Pestov NA, Kulaeva OI, Clark DJ, Studitsky VM. Transcription-induced DNA supercoiling: New roles of intranucleosomal DNA loops in DNA repair and transcription. Transcription. 2016;7(3):91-5. Epub 2016/04/27. doi: 10.1080/21541264.2016.1182240. PubMed PMID: 27115204; PMCID: PMC4984681.
Chang HW, Pandey M, Kulaeva OI, Patel SS, Studitsky VM. Overcoming a nucleosomal barrier to replication. Science advances. 2016;2(11):e1601865. Epub 2016/11/17. doi: 10.1126/sciadv.1601865. PubMed PMID: 27847876; PMCID: PMC5106197.
This Fox Chase professor participates in the Undergraduate Summer Research Fellowship.
Learn more about Research Volunteering.