This Fox Chase professor participates in the Undergraduate Summer Research Fellowship.
Learn more about Research Volunteering.

This Fox Chase professor participates in the Undergraduate Summer Research Fellowship.
Learn more about Research Volunteering.
Assistant Research Professor
Adjunct Professor, Department of Medical Genetics and Molecular Biochemistry, Lewis Katz School of Medicine, Temple University
My lab studies the early events of retroviral infection, with a focus on the essential viral integrase protein that inserts viral DNA into the host genome. Integrase is a validated target for antiviral therapeutics including those that target HIV-AIDS and potentially other retroviral-induced cancers. One project in the lab is centered on the host proteins that associate with the pre- integration complex (PIC), including both factors that are co-opted by the virus to promote replication and those that represent the host cell’s innate immune response to incoming viral infection. We are interested in discerning the molecular interactions of the host restriction factor Daxx with the integrase protein of the PIC, and how that results in silencing of proviral expression.
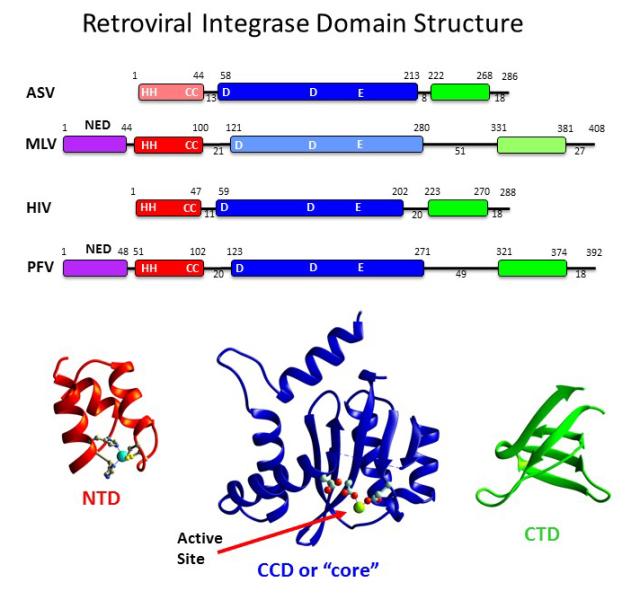
Secondly, our major focus has been to understand the biochemistry, enzymology, multimerization and structure of retroviral integrases. We have utilized a variety of biophysical methods to characterize the structure of integrases, including small angle X-ray scattering, X- ray crystallography, catalytic assays, and methods to detect multimerization, and integrase-DNA binding. Currently we are using computational methods to engineer an HIV-integrase that forms stable complexes with viral DNA (called intasomes) that are amenable to crystallography trials and structure determination. The absence of an atomic resolution description of the HIV intasome is a major deficit in the development of anti-HIV therapeutics. In addition, we are currently screening for small molecule allosteric inhibitors that interfere with integrase multimerization as a new strategy for anti- retroviral therapeutics.
Lastly, in managing the Fox Chase Molecular Modeling Facility, I have enjoyed being a part of a collaborative team environment for translational medicine. The facility has contributed to important papers involving epigenetics and mitosis, cell signaling, kinase inhibition in cancer therapeutics, and nucleosome structure and regulation. In addition to collaborating with laboratory investigators, more recently I have been assisting clinical investigators in interpreting the results of sequencing of tumor samples and germline DNA by determining the likely impact of missense mutations on protein function and cellular processes. With the advent of incorporating tumor gene sequencing technologies to the diagnostic repertoire of personalized cancer care, there is a pressing need to understand the structural and biochemical effects of the mutations that are found. Since 2011, we have undertaken projects with 15 investigators dealing with the structural and functional consequences of missense mutations identified in breast, kidney, prostate, bladder, lung, genitourinary, colon and other GI cancers.


Structure and biochemistry of retroviral integrases and other early events of retroviral infection
Kueppers F., Andrake M.D., Xu Q., Dunbrack R.L., Jr., Kim J. ,Sanders C.L., Protein modeling to assess the pathogenicity of rare variants of serpina1 in patients suspected of having alpha 1 antitrypsin deficiency. BMC Med Genet. 20(1): 125, 2019. PMC6631921. 1.740
Gupta S., Kelow S., Wang L., Andrake M., Dunbrack R.L., Jr. ,Kruger W.D., Mouse modeling and structural analysis of the p.G307s mutation in human cystathionine beta-synthase (cbs) reveal effects on cbs activity but not stability. J Biol Chem. 293(36): 13921-13931, 2018. PMC6130948. 4.106
Bojja, R.S., Andrake, M.D., Merkel, G., Weigand, S., Dunbrack, R.L., Jr., Skalka, A.M. Architecture and assembly of HIV integrase multimers in the absence of DNA substrates. J. Biol. Chem. 288:7373-7386, 2013. PubMed
Peletskaya, E., Andrake, M., Gustchina, A., Merkel, G., Alexandratos, J., Zhou, D., Bojja, R.S., Satoh, T., Potapov, M., Kogon, A., Potapov, V., Wlodawer, A., Skalka, A.M. Localization of ASV integrase-DNA contacts by site-directed crosslinking and their structural analysis. PLoS One 6:e27751, 2011. PubMed
Katz, R.A., Merkel, G., Andrake, M.D., Roder, H., Skalka, A.M. Retroviral integrases promote fraying of viral DNA ends. J. Biol. Chem. 286:25710-25718, 2011. PubMed
Bojja, R.S., Andrake, M.D., Weigand, S., Merkel, G., Yarychkivska, O., Henderson, A., Kummerling, M., Skalka, A.M. Architecture of a full-length retroviral integrase monomer and dimer, revealed by small angle X-ray scattering and chemical cross-linking. J. Biol. Chem. 286:17047-17059, 2011. PubMed
Andrake, M.D., Ramcharan, J., Merkel, G., Zhao, X.Z., Burke, T.R., Jr., Skalka, A.M. Comparison of metal-dependent catalysis by HIV-1 and ASV integrase proteins using a new and rapid, moderate throughput assay for joining activity in solution. AIDS Res. Ther.6:14, 2009. PubMed
Merkel, G., Andrake, M.D., Ramcharan, J., Skalka, A.M. Oligonucleotide-based assays for integrase activity. Methods 47:243-248, 2009. PubMed
Andrake, M.D., Sauter, M.M., Boland, K., Goldstein, A.D., Hussein, M., Skalka, A.M. Nuclear import of Avian Sarcoma Virus integrase is facilitated by host cell factors. Retrovirology 5:73, 2008. PubMed
Andrake MD, Skalka AM. Retroviral integrase, putting the pieces together. J Biol Chem. 1996 Aug 16;271(33):19633-6. PubMed.
Andrake MD, Skalka AM. Multimerization determinants reside in both the catalytic core and C terminus of avian sarcoma virus integrase. J Biol Chem. 1995 Dec 8;270(49):29299-306. PubMed
This Fox Chase professor participates in the Undergraduate Summer Research Fellowship.
Learn more about Research Volunteering.