This Fox Chase professor participates in the Undergraduate Summer Research Fellowship.
Learn more about Research Volunteering.

This Fox Chase professor participates in the Undergraduate Summer Research Fellowship.
Learn more about Research Volunteering.
Professor
Professor, Department of Biochemistry, Temple University
Adjunct Professor, Department of Biochemistry and Biophysics, University of Pennsylvania
Hong Cheng, PhD
Staff Scientist
Ruzaliya Fazlieva, MS
Scientific Technician II
Ming Xu, Ph.D.
Postdoctoral Associate
Protein folding, structure and function
Our research is aimed at understanding fundamental questions in protein science, including mechanisms of protein folding, protein structure-function relationships, and the impact of protein dynamics and intrinsic disorder on protein function. My group approaches these problems using various biophysical tools, including NMR, fluorescence and other optical techniques, rapid kinetics and protein engineering.
A central theme of our research concerns the early stages of protein folding, which are critical for understanding how the native structure of a protein, and ultimately its function, are encoded in the amino acid sequence. The insight gained not only provides a basis for protein structure prediction and de novo design, but also contributes to our mechanistic understanding and treatment of a wide range of diseases that involve aggregation of denatured or misfolded proteins. The stability and folding dynamics of proteins also have major implications with respect to understanding of the physiological consequences of mutations, in vivo folding and other cellular processes, such as trafficking and degradation. We study the dynamics of protein folding on a microsecond time scale by coupling advanced mixing techniques with various detection methods, including fluorescence and H/D exchange labeling experiments with NMR detection. In combination with protein engineering, these approaches have provided detailed insight into the folding mechanisms of a diverse set of proteins.
A recent effort has been to elucidate the mechanism of coupled protein folding/ligand binding reactions. We explored the ligand-induced folding reaction of staphylococcal nuclease (SNase) under conditions (2.9 M Urea) where the free protein is unfolded, but the complex with a nucleotide analog (prAp) is folded, using optically monitored kinetic measurements, NMR and molecular dynamics simulations. Theoretical modeling of the kinetic data using statistical mechanics showed that the conformational changes precede binding at low ligand concentrations (“conformational selection”), whereas the second-order binding step preceded formation of the native structure at high ligand concentrations (“induced fit”). Real-time 1D and 2D NMR measurements confirmed that an encounter complex of prAp with a partially structured, but dynamic, form of SNase accumulates transiently at high ligand concentrations. The results shed new light on the structural/energetic basis of coupled folding/binding reactions, which are highly relevant not only for understanding molecular recognition in cases where at least one of the binding partners is intrinsically disordered.
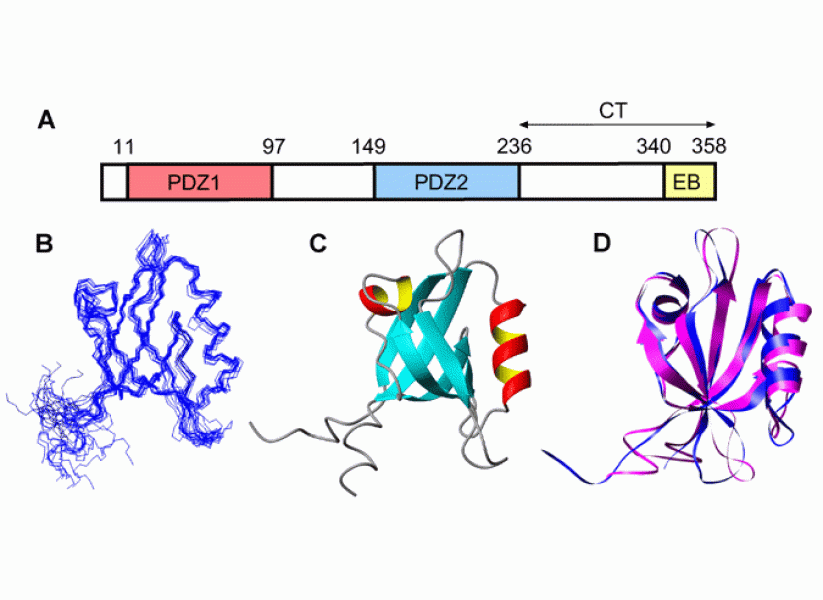
Our group also investigates the structure, dynamics and molecular interactions of various proteins of biomedical interest in solution by using NMR spectroscopy and other biophysical methods. An ongoing project is aimed at understanding the structural and dynamic properties of Na+/H+ exchanger regulatory factor (NHERF), a signaling protein comprising two PDZ domains and a C-terminal ezrin binding motif, as well as two long intrinsically disordered regions. Detailed NMR and thermodynamic studies have shown that the activity of NHERF as a signaling adaptor at the membrane-cytoskeleton interface is regulated by a complex equilibrium between various open and closed conformations. By combining our biophysical approaches with cellular imaging and flow cytometry, we have gained detailed insight into the mechanism by which NHERF regulates the signaling activity and trafficking of the epidermal growth factor receptor (EGFR).
More broadly, we are interested in understanding how interactions between multivalent binding partners, such as NHERF or the SH2-domain containing phosphatase Shp2, and intrinsically disordered cytosolic regions of membrane receptors, such as EGFR or the killer cell inhibitory receptor KIR3DL, can mediate receptor clustering and phase separation on the plasma membrane. To resolve and assign the crowded NMR spectra of these intrinsically disordered protein regions (IDRs) we make extensive use of 13C-detected NMR techniques, including a novel technique for mapping recognition sites for globular binding partners on IDRs. By applying these approaches to the cytosolic tail of KIR3DL1, we gained detailed insight into the secondary structure propensities of this 84-residue IDR, membrane-interacting regions, and binding sites for a key interaction partner, the phosphatase Shp2, which recognizes a pair of phospho-tyrosine sequence motifs via a pair of SH2 domains. These findings provide a structural basis for understanding how interactions between two bivalent binding partners regulate receptor signaling.
This Fox Chase professor participates in the Undergraduate Summer Research Fellowship.
Learn more about Research Volunteering.