This Fox Chase professor participates in the Undergraduate Summer Research Fellowship.
Learn more about Research Volunteering.

This Fox Chase professor participates in the Undergraduate Summer Research Fellowship.
Learn more about Research Volunteering.

We are interested in the biology of human pathogenic viruses with an emphasis on mechanisms of viral replication and host-virus interactions that play a role in innate immunity. Our investigations on hepatitis B virus (HBV) lead to the identification of the signals required for reverse transcription of the viral DNA and provided the basis for the current model for hepadnavirus replication. We discovered that the hepatitis B polymerase could be expressed in enzymatically active form in the presence of the heat shock protein 90 complex. Moreover, we demonstrated that recovery from chronic hepatitis B infections requires massive destruction of infected hepatocytes.
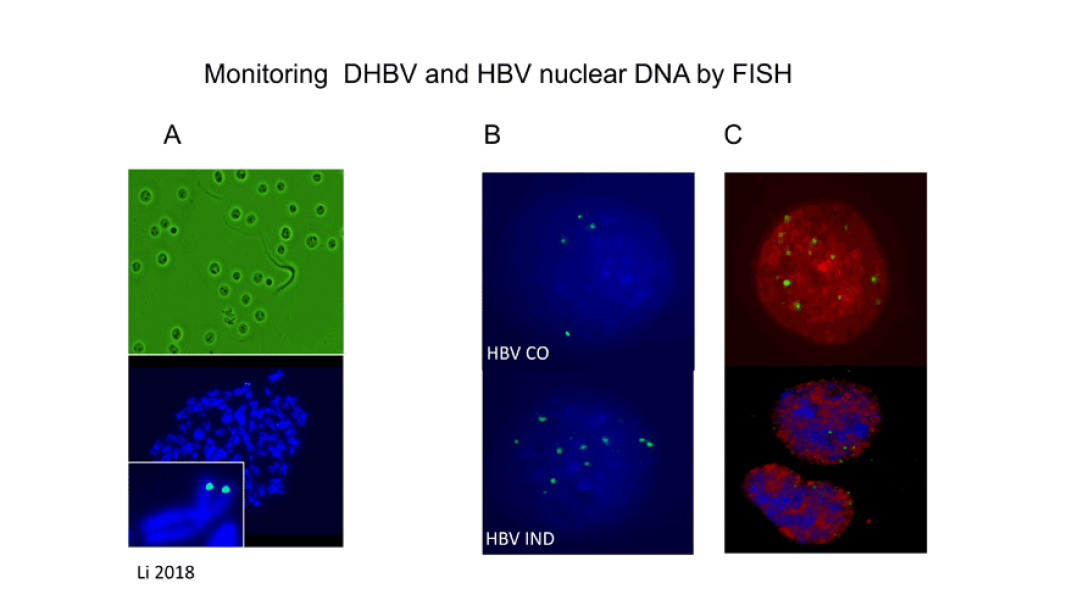
In line with our interest in HBV biology, the goal of our current research effort is to investigate one of the least understood steps in HBV replication: the mechanism by which the viral genome is converted into a covalently closed circular (ccc) DNA form and how intracellular amplification of cccDNA is regulated. To conduct our studies, we have developed a CRISPR/Cas9 platform permitting HBV infection of cells with specific gene knockouts. In addition, we are exploring a novel strategy to eliminate cccDNA from infected cells with the help of the CRISPR/Cas9 system. Finally, we are developing methods to visualize individual copies of cccDNA in HBV infected cells. We seek to determine the localization of cccDNA in nuclei of infected cells and to monitor the fate of cccDNA under conditions mimicking natural recovery from acute HBV infections. Our long-term goal is to provide novel insights into the biology of cccDNA that can be used for the development of therapies to cure chronic hepatitis B.
Li M, Sohn JA, Seeger C. Distribution of Hepatitis B Virus Nuclear DNA. J Virol, 92(1), 2018. PMC5730781
Seeger, C and Sohn, J.A. Complete spectrum of CRISPR/Cas9-induced mutations on HBV cccDNA. Molecular Therapy, 24:1258-66, 2016. https://www.ncbi.nlm.nih.gov/pubmed/27203444
Seeger, C and Sohn J.A. Targeting HBV cccDNA with CRISPR/Cas9. Molecular Therapy-Nucleic Acids, e216; doi:10.1038/mtna.2014.68, 2014. PubMed
Sohn, J.A., Litwin, S., Seeger, C. Mechanism for CCC DNA synthesis in hepadnaviruses. PLoS ONE 4(11): e8093, 2009.
Hu, J., Seeger, C. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc. Natl. Acad. Sci. USA 93:1060-1064, 1996. https://www.ncbi.nlm.nih.gov/pubmed/8577714
Wang, G.-H., Seeger, C. The reverse transcriptase of hepatitis B virus acts as a protein-primer for viral DNA synthesis. Cell 71:663-670, 1992. https://www.ncbi.nlm.nih.gov/pubmed/1384989
Seeger, C., Ganem, D., Varmus, H.E. Biochemical and genetic evidence for the hepatitis B virus replication strategy. Science 232:477‑487, 1986. https://www.ncbi.nlm.nih.gov/pubmed/3961490
This Fox Chase professor participates in the Undergraduate Summer Research Fellowship.
Learn more about Research Volunteering.