This Fox Chase professor participates in the Undergraduate Summer Research Fellowship.
Learn more about Research Volunteering.
Related Articles
00 / 00
-

Dr. Whetstine
-

Dr Whetstine at 2023 Paws for the Cause
-

Dr. Whetstine
-

Dr. Johnathan R. Whetstine, director of the Cancer Epigenetics Institute at Fox Chase
-

Jaye Gardiner, PhD, a postdoctoral fellow at Fox Chase Cancer Center
-

-

-

-

-

-

-

-

-

A photograph of two Fox Chase doctors sitting in a dim room, one gesturing at a screen with various blue splotches displayed.
-

-

The American Lung Association’s Lung Cancer Discovery Award, a $100,000 grant to research epigenetic factors affecting lung cancer.
-

-

The support of our friends and donors who have made these endowments possible gives us the freedom to pursue yet-uncharted areas.

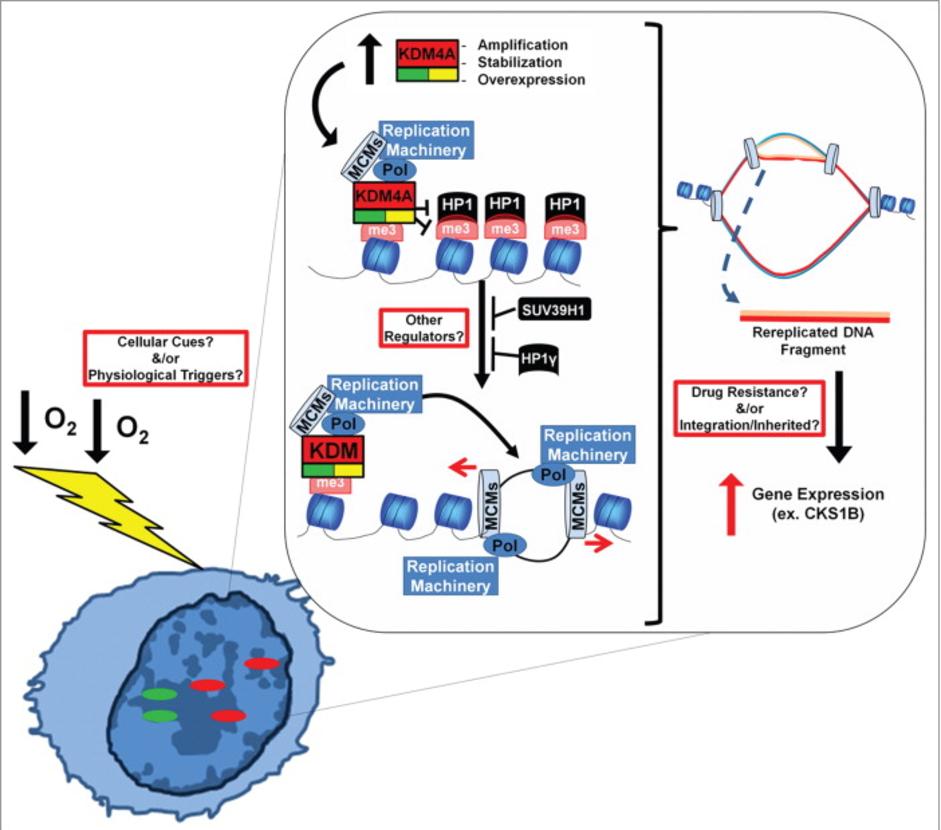
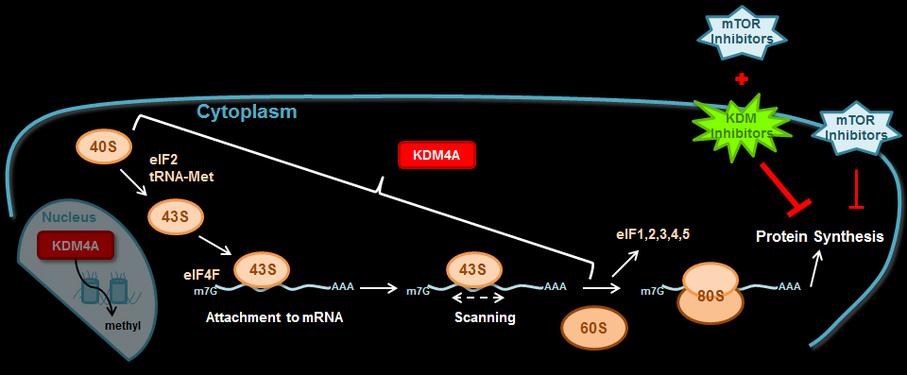
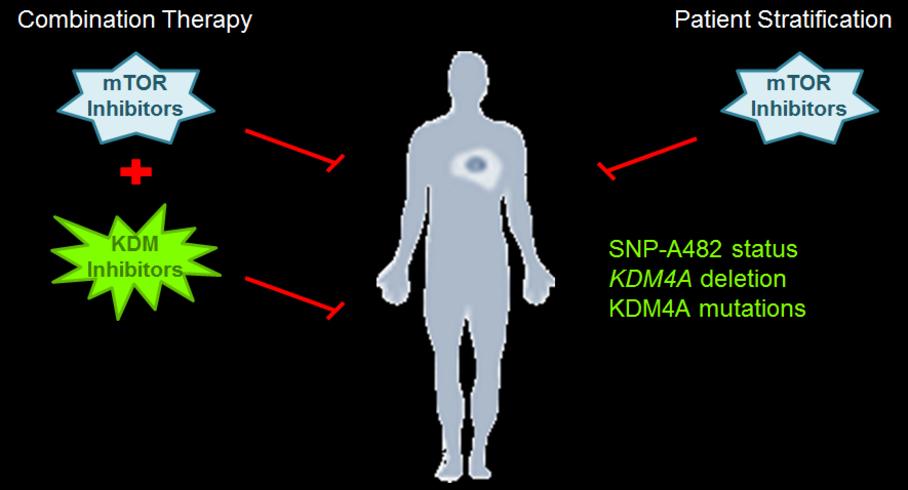
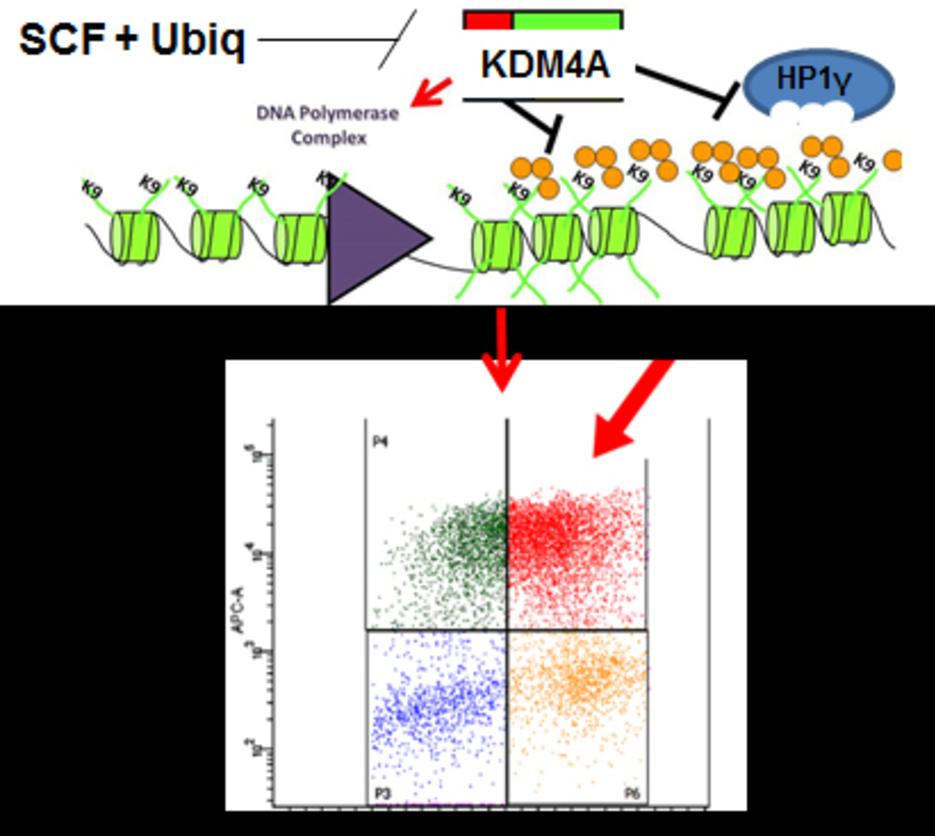





















Patient comments